Trapping a plastic-degrading enzyme in a synthetic nanocage protects it from breaking down under heat or in organic solvents. While the caged enzyme retains all of its catalytic activity, it withstands temperatures much hotter than its usual melting point and becomes 1000 times more resistant to solvents. Similar nanocages could protect and extend the life of protein-based drugs, vaccines or diagnostics.
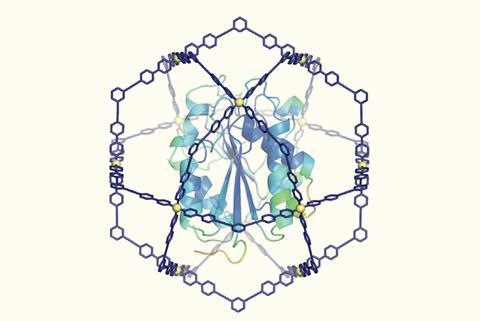
In 2004 , Makoto Fujita and his group at the University of Tokyo, Japan, prepared a cage molecule from 12 palladium ions and 24 bridging ligands. The components self-assembled into a large spherical framework with a diameter of approximately 7nm. ‘Chemists had never synthesised such a huge cage, nor envisioned fully encapsulating therein a giant guest molecule like a protein,’ explains Makoto Fujita.
Now, a team around Makoto Fujita and his colleague Daishi Fujita have used their giant cage as a protein protector. A formyl pyridine group in the cage ligand interacts with proteins’ N-terminus and lysine side chains. The ligands pre-assemble around the protein, and palladium is added as a clamp to the close the cage.
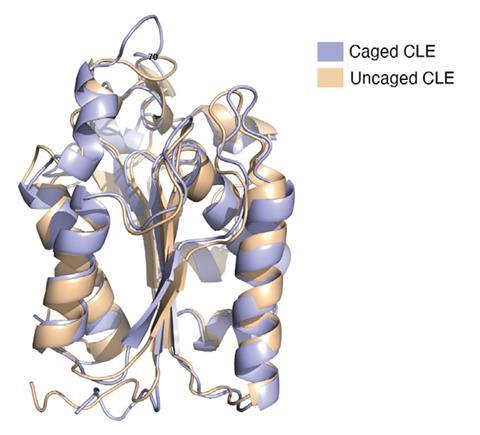
The team tested their cage on a cutinase, a 21kDA 206 amino acids enzyme that is known to degrade plastic. With a maximum diameter of about 4.5nm, the protein fits nicely inside the cage. When the confined enzyme was exposed to an organic solvent, its half-life was prolonged 1,000-fold compared with the uncaged version. Moreover, the caged enzyme did not melt when heated above 100°C, although the uncaged enzyme’s melting point is around 50°C .
‘Enzymes confined in a well-fitted space allow the molecular movements necessary for enzymatic reactions but inhibit the major structural changes that lead to denaturation,’ explains Daishi Fujita. The Japanese team hopes to use their cage-reinforced enzyme to degrade hydrophobic plastics on an industrial scale.
‘Putting enzymes into a confined space is not new,’ says notes polymer chemist Martina Stenzel at the University of New South Wales in Australia. Other strategies to trap proteins include metal–organic frameworks, single enzyme nanoparticles, metallic gold and mesoporous silica. However, many ‘are based on polymers, which are mobile and can to some extent adjust to the movement of the enzyme’, Stenzel says. Here, the researchers used a hard structure, which fits around the enzyme and constrains movement further, she explains. ‘As a result, the stability seen here is very impressive.’
‘Stopping denaturation can prolong the life-time of the enzyme, [and] therefore can have economic huge benefits,’ Stenzel notes. Storage challenges with enzymes, such as the need for temperature control, can render enzyme-based diagnostic tests unreliable – cages could protect them, she adds.
‘Entrapment of enzymes in these large self-assembled molecular cages, leading to well defined structures, is a remarkable achievement,’ comments Joost Reek, supramolecular chemist at the University of Amsterdam in the Netherlands. Preparing cages with different symmetries may be worth pursuing, he notes.
However, ‘since molecular self-assembly typically gives spherical products, preparing a cage for proteins with elongated shapes, for example, is another technological challenge,’ Daishi Fujita explains. For now, proteins larger than 10 nm remain too bulky for encapsulation.
Nevertheless, considering the enzymatic activity and improved stability, ‘the current caged protein would be pretty promising for industrial biocatalyst applications’, says Hai-Bo Yang, a supramolecular chemist at East China Normal University.