# **Generative AI Boosts Discovery of Antimicrobial Peptides to Fight Drug-Resistant Infections**
Antibiotic resistance ranks among the most pressing global health issues, with projections indicating that drug-resistant infections could outpace cancer as the leading cause of death by 2050. Addressing this urgent challenge, a research team under the leadership of Wenqiang Chang at Shandong University, China, has created a groundbreaking generative AI model that can swiftly design a wide array of antimicrobial peptide (AMP) structures. This innovation carries the potential to transform drug discovery by pinpointing effective treatments for infections caused by resistant pathogens.
## **Utilizing Artificial Intelligence in Drug Discovery**
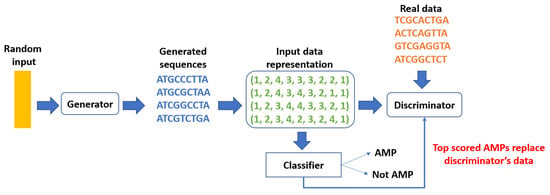
Antimicrobial peptides (AMPs) have emerged as a viable alternative to traditional antibiotics, given their capacity to target bacterial membranes—a mechanism that complicates the bacteria’s ability to develop resistance. Nevertheless, the extensive variety of potential AMP structures poses a significant barrier to the identification of candidates with robust antimicrobial properties. Conventional AI methods aimed at generating new peptide sequences frequently face challenges regarding diversity, yielding results that closely mimic their training data.
To overcome this limitation, Chang’s team employed a **latent diffusion model**, a generative AI system designed to function within compressed data formats. This model generated an extensive library of **2.8 million peptides**, encoding them in a numerical framework referred to as *latent space*. The system then applied controlled signal noise to the dataset before methodically denoising the information to derive unique and varied AMP structures.
> *“The denoising process is stochastic, meaning the model does not always remove noise in the same manner. This slight randomness in denoising pathways ensures diversity in the produced peptides,”* states Jon Stokes, an antimicrobial chemical biologist at McMaster University, who was not part of the research.
## **Experimental Validation: Discovering Lead Candidates**
After the generative AI system generated a collection of **600,000 candidate sequences**, the peptides underwent filtering through a classification pipeline that pinpointed those most likely to demonstrate antimicrobial properties. From this selection, the team **experimentally evaluated 40 peptides** against various fungal and bacterial pathogens. Remarkably, **25 out of the 40 showcased antimicrobial effects**, indicating a high success ratio.
To further evaluate safety and efficacy, the researchers conducted *in vivo* examinations, leading to two distinguished candidates: **AMP-24 (with antibacterial effects) and AMP-29 (with antifungal effects)**. These peptides were successfully tested in mice, indicating their potential as effective treatments for microbial infections.
## **Broadening the Possibilities of AI-Driven Drug Discovery**
The achievements of Chang’s model illustrate how generative AI can expedite drug development by producing diverse and effective compounds on an unprecedented scale. Stokes, who has expertise in AI-assisted antimicrobial discovery, emphasizes the importance of this advancement.
> *“Their ability to generate peptides that demonstrate *in vivo* potential and have acceptable toxicity profiles is incredibly promising,”* he notes. *“I believe there will be a wealth of valuable insights derived from this.”*
However, Stokes highlights the necessity for ongoing enhancements, recommending that **toxicity assessments should be integrated earlier** within the AI workflow to facilitate the identification of clinically viable candidates.
## **Future Directions: Clinical Translation and Wider Applications**
Chang’s team is already advancing **AMP-24** toward clinical development while refining their AI model to optimize several parameters, including cytotoxicity. Besides AMPs, they foresee the application of their methodology across multiple facets of drug discovery.
> *“The fundamental value of [the model] lies in expediting drug discovery [and] reducing development expenses, potentially revolutionizing traditional molecular discovery methods,”* Chang asserts.
This breakthrough emphasizes the potential of AI in transforming the future of medicine. By facilitating the rapid generation and assessment of bioactive molecules, AI-driven drug discovery models possess the capability to **counter antibiotic resistance, lower development costs, and unveil new treatment pathways** for a range of diseases.
As researchers continue to refine this methodology and broaden its applications, we may soon observe AI taking a pivotal role in **accelerating the development of next-generation antimicrobial therapies**, providing hope in the battle against drug-resistant infections.